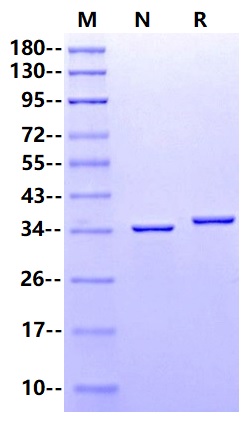
35.7kDa (Reducing)
>95% by SDS-PAGE
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, 50% Glycerol, pH7.5
· 12 months from date of receipt, -20 to -70 °C as supplied.
· Please avoid repeated freeze-thaw cycles.
1. Frank Maley and Robert B. Trimble and Anthony L. Tarentino and Thomas H. Plummer Jr. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases[J]. Analytical Biochemistry, 1989.
Peptide: N-glycosidase F (PNGase F) is an asparagine amidase produced by Flavobacterium meningosept-icum that serves as a useful tool in the research on protein N-glycosylation. Recombinant expression in E.coli. The cleavage site of PNGase F is the amide bond between N-acetylglucosamine (GlcNAc) and aspartate residues on the medial side of the glycoprotein, and converts aspartyl to aspartic acid on the enzymolysis protein. This product is often used for complete deglycosylation of antibodies and their associated proteins.
The PNGase F Glycan Cleavage Kit includes all components necessary to perform the enzymatic removal of almost all N-linked oligosaccharides from glycoproteins. The kit includes recombinant Peptide N-Glycosidase F(PNGase F) enzyme, which cleaves N-glycan chains at the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides, and a 10X reaction buffer.
Components | Amount |
PNGase F* | 40000U/ml |
Reaction Buffer (10×) | 200mM Tris, PH 7.5 25°C |
Denaturing Buffer(10×) | 5% SDS、400 mM DTT |
NP-40(10×) | 10% NP-40 in MilliQ-H2O |
* One unit is defined as the amount of enzyme required to remove > 95% of the carbohydrate from 10 μg of denatured RNase B in 1 hour at 37°C in a total reaction volume of 10 μl.
一. Denaturing Reaction Conditions:
1. Combine 1-20 µg of glycoprotein, 1 µl of Denaturing Buffer (10X) and H2O (if necessary) to make a 10 µl total reaction volume.
2. Denaturation is terminated by heating to 100℃ for 10-20min and cooling to room temperature.
3. Make a total reaction volume of 20 µl by adding 2 µl Reaction Buffer (10×), 2 µl 10% NP-40 and 6 µl H2O.
4. Add 1 µl PNGase F, mix gently.
5. Enzymatic digestion at 37℃ for 1h
6. Analyze by the method of SDS-PAGE
二. Non-Denaturing Reaction Conditions:
1. Combine 1-20 µg of glycoprotein, 2 µl of Reaction Buffer (10×) and H2O (if necessary) to make a 20 µl total reaction volume.
2. Add 2-5 µl PNGase F, mix gently.
3. Enzymatic digestion at 37°C for 4 - 24 hours.
4. Analyze by the method of SDS-PAGE
1.The target protein should be in a solution compatible with Rapid PNGase F activity. Avoid buffers containing SDS, as it inhibits PNGase F. Common stabilizing reagents such as Tween, Triton X-100, NP-40, octyl glucoside and non-detergent sulfobetaine, as well as traces of organic solvents, can prevent optimal rapid deglycosylation.
2.N-linked glycans containing core α1-3 Fucose are not cleaved by PNGase F.
3.For deglycosylation of native glycoprotiens, increased incubation time and increased amount of enzyme may be needed.
4.Optimize reaction conditions for each substrate.
5.If a larger amount of glycoprotein is used, scale up reaction volumes accordingly.
6.Although this product has been optimized for the rapid removal of N-glycans from antibodies, it can be utilized with various other glycoproteins.
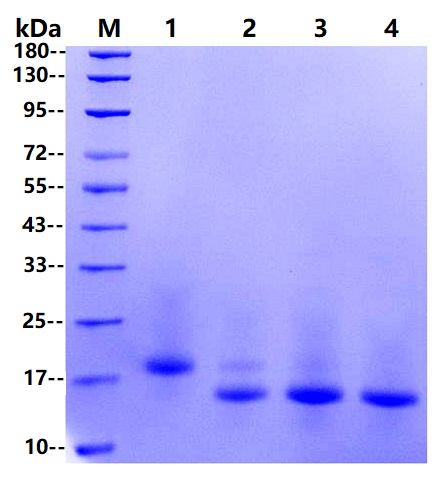
M marker
1: Rnase B 10μg
2: Rnase B 10μg+0.25U PNGF
3: Rnase B 10μg+0.5U PNGF
4: Rnase B 10μg+1U PNGF