64.9kDa (Reducing)
Store at -25 ~ -15℃
for 2 years
[1] Zhu Z, Samuelson J C, Zhou J, et al.Engineering Strand-specific DNA Nicking Enzymes from the Type IIS Restriction Endonucleases BsaI, BsmBI, and BsmAI[J].Journal of Molecular Biology, 2004, 337(3):573-583.
[2] Lee J H, Won H J, Oh E S,etal. Golden Gate
Cloning-Compatible DNA Replicon/2A-Mediated Polycistronic Vectors for
Plants[J]. Frontiers in Plant Science, 2020, 11.
BsaI is a Type IIs restriction enzyme that can recognize non-palindromic sequences and cut outside of the recognition sequence. It is commonly used for Golden Gate assembly and enzymatic cleavage of plasmids to prepare linear DNA fragments with poly (A/T/G/C) endings and obtain specific sticky ends.
Recognition site:
5'-GGTCTC(N)1↓-3'
3'-CCAGAG(N)5↑-5'
Storage Solution: 20 U/μl BsaⅠ、10 mM Tris-HCl、300 mM NaCl、1 mM DTT、0.1 mM EDTA、500 µg/ml BSA、50% Glycerol(pH 7.4 @ 25°C)
10*Reaction Buffer:500 mM Potassium Acetate、200 mM Tris-acetate、100 mM Magnesium Acetate、1mg/ml BSA(pH 7.4 @ 25°C)
This step is suitable for linearization of 1 μg DNA (≥100 nt) and can be scaled up according to experimental needs.
1)Add the following components in sequence
| Components |
Volume
|
Plasmid DNA
|
1μg DNA
|
10*Reaction Buffer
|
5μl |
BsaⅠ (20 U/μl)
|
1μl
|
RNase-free ddH2O
| Up to 50μl |
2)Incubate at 37°C 1h
3)DNA linearization is complete, and subsequent experiments can be performed.This step is suitable for linearization of 1 μg DNA (≥100 nt) and can be scaled up according to experimental needs.
1. star activity may result from a glycerol concentration of >5%; 2. Please avoid repeated freeze-thaw cycles.
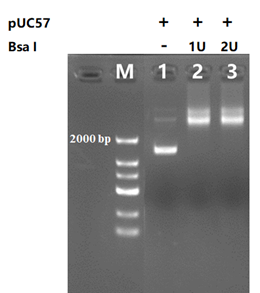
The results of 1μg pUC57 plasmid digestion separated under different quantity of BsaⅠ, The reaction was incubated for 60 minutes at 37°C, and 1% agarose gel was used for electrophoresis analysis after reaction.
M, marker;
Lane 1 1μg pUC57;
Lane 2 1μg pUC57 add 1U BsaⅠ
Lane 3 1μg pUC57 add 2U BsaⅠ
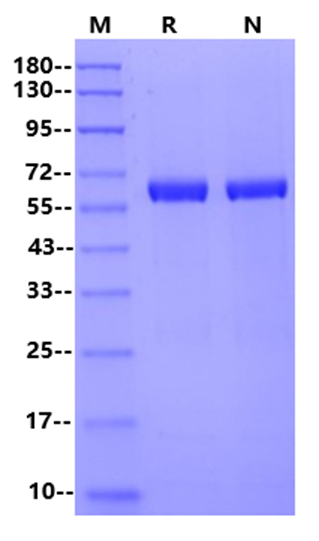
1μg (R: reducing
condition, N: non-reducing condition).